TP4
From RAS mutations in Noonan syndrome to interaction network profiling and mechanism-based target identification
This subproject focuses on the RAS-MAPK interaction networks in order to explore new molecular and cellular control mechanisms and to identify potential novel RASopathy-relevant targets. Therefore, it was mandatory to establish new and to further develop existing methods, and to optimize culturing of the cell lines of interests, including iPSCs derived from patients with RASopathies and their cardiomyocyte derivatives. A direct approach to identify novel proteins as integral elements of the RAS-MAPK interaction networks, such as accessory/scaffold proteins and/or with lipid membrane, is the analysis RAS mutations in codons 5, 50, 58, 71 and 153 identified in individuals with Noonan syndrome. These RAS variants did not exhibit major alterations using established biochemical methods under cell-free conditions while being hyperactive in cells. Another approach is identifying, evaluating and characterizing novel components of the RAS-RAF1 complex as well as novel RAF1 substrates in three independent ways: (1) Detergent-free fractionation combined with 2D blue native PAGE; (2) LC/MS, quantitative SILAC phosphoproteomics; (3) Cell-free liposome reconstitution of RAS signaling. Functional characterization of newly identified and validated proteins will be crucial to understand the pathogenic mechanisms underlying RASopathy. In addition, we will develop selective cell-based fluorescent probes, which enables us to both monitor in real-time the activity of downstream signaling pathways of RAS and quantitatively measure the activities and specificity of various small molecule inhibitors. These investigations will ultimately disclose further molecular details of dysregulated signaling pathways.
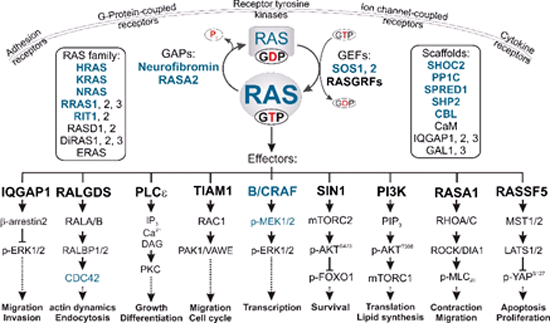
Multiple signal transduction pathways downstream of RAS. The binding of GTP to RAS proteins enables them to undergo high affinity interactions with multiple effectors, which links different extracellular cues to distinct downstream pathways. GEFs and GAPs tightly control two intrinsic reactions, nucleotide exchange and GTP hydrolysis. In a polarized cell, RAS activation and signaling can be accomplished both at different sites of the plasma membrane and in endomembrane compartments. Different members of the RAS family and potential scaffold proteins, potentiating or attenuating signal transduction, are shown in boxes. RASopathy-associated proteins are shown in blue.
Major goals:
- Understanding the RAS interaction networks and identifying and validating new RAS associating proteins as potential new drug targets
- Understanding the impact of the non-conventional RAS variants on the RAS interaction networks
- Investigating membrane interaction, regulation and signaling of RAS using an in vitro reconstitution model of prenylated RAS proteins on synthetic liposomes
- Establishing fluorescent probes to monitor in real-time RAS-RAF-MEK-ERK and RAS-PI3K-AKT pathways and to decipher critical control mechanisms
Publications:
Altmüller F, Pothula S, Annamneedi A, Nakhaei-Rad S, Montenegro-Venegas C, Pina-Fernández E, Marini C, Santos M, Schanze D, Montag D, Ahmadian MR, Stork O, Zenker M, Fejtova A. Aberrant neuronal activity-induced signaling and gene expression in a mouse model of RASopathy. PLoS Genet. 2017 Mar 27;13(3):e1006684.
PMID: 28346493
Martinelli S, Krumbach OHF, Pantaleoni F, Coppola S, Amin E, Pannone L, Nouri K, Farina L, Dvorsky R, Lepri F, Buchholzer M, Konopatzki R, Walsh L, Payne K, Pierpont ME, Vergano SS, Langley KG, Larsen D, Farwell KD, Tang S, Mroske C, Gallotta I, Di Schiavi E, Della Monica M, Lugli L, Rossi C, Seri M, Cocchi G, Henderson L, Baskin B, Alders M, Mendoza-Londono R, Dupuis L, Nickerson DA, Chong JX; University of Washington Center for Mendelian Genomics, Meeks N, Brown K, Causey T, Cho MT, Demuth S, Digilio MC, Gelb BD, Bamshad MJ, Zenker M, Ahmadian MR, Hennekam RC, Tartaglia M, Mirzaa GM. Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes. Am J Hum Genet. 2018 Jan 17. PMID: 29394990
Haghighi F, Dahlmann J, Nakhaei-Rad S, Lang A, Kutschka I, Zenker M, Kensah G, Piekorz RP, Ahmadian MR. bFGF-mediated pluripotency maintenance in human induced pluripotent stem cells is associated with NRAS-MAPK signaling. Cell Commun Signal 2018; 16:96. PMID: 30518391
Motta M, Sagi-Dain L, Krumbach OHF, Hahn A, Peleg A, German A, Lissewski C, Coppola S, Pantaleoni F, Kocherscheid L, Altmüller F, Schanze D, Logeswaran T, Chahrokh-Zadeh S, Munzig A, Nakhaei-Rad S, Cavé H, Ahmadian MR, Tartaglia M, Zenker M. Activating MRAS mutations cause Noonan syndrome associated with hypertrophic cardiomyopathy. Hum Mol Genet 2019 May 21. [Epub ahead of print] PMID: 31108500
Capri Y, Flex E, Krumbach OHF, Carpentieri G, Cecchetti S, Lißewski C, Rezaei Adariani S, Schanze D, Brinkmann J, Piard J, Pantaleoni F, Lepri FR, Goh ES, Chong K, Stieglitz E, Meyer J, Kuechler A, Bramswig NC, Sacharow S, Strullu M, Vial Y, Vignal C, Kensah G, Cuturilo G, Kazemein Jasemi NS, Dvorsky R, Monaghan KG, Vincent LM, Cavé H, Verloes A, Ahmadian MR, Tartaglia M, Zenker M. Activating Mutations of RRAS2 Are a Rare Cause of Noonan Syndrome. Am J Hum Genet 2019; 104:1223-1232. PMID: 31130282
TP4 project description - previous funding period FP1








